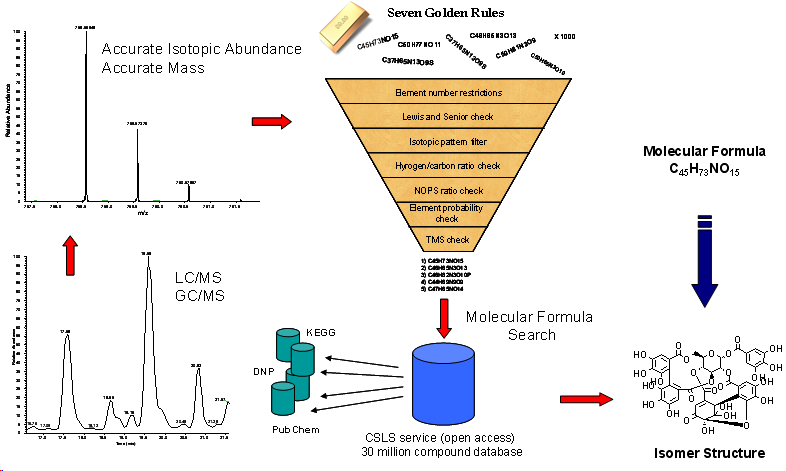
The Seven Golden Rules provide a list of highly probable formulas. For that reason the EXCEL script contains a large list of 750,000 existing molecular formulas covering more than 10 million existing compounds. Additionally the 7GR are connected via hyperlinks to the current largest open access database for small molecules the Chemical Structure Lookup Service (CSLS). This meta-database contains links (pointers) to PubChem, DNP, Wombat and other free and commercial chemical databases. The CSLS covers 39 million indexed structures from over 80 databases (27 million unique structures) and is currently the largest semi open-access database of this kind. For every molecular formula hit the corresponding structural isomers can be found. Example search for C41H64O13.
As presented already in the publication; the Seven Golden Rules are highly effective if molecular formula target databases are used. Such databases can be toxic compound databases (TSCA), natural compound databases (DNP, KEGG, LipidMaps) or drug databases (MDL DDR, RedPoll Drugbank or PubChem itself. In such a case the correct molecular formula is found with a high probability (usually 80-99%. But such annotations should not be confused with unambiguous identifications. Any true identification of a structural isomer from a molecular formula needs additional steps (like mass spectral fragmentations, matching physico-chemical properties, retention indices, NMR) or confirmation by an external standard compound.
Why not directly use CAS or Beilstein? CAS and Beilstein have the largest size and best data quality of all chemical databases, because they are curated. They have the large advance to cover most of the chemical literature or sometimes link directly to the PDF of the publication where a structure was mentioned. CAS and Beilstein also were included during the development of the Seven Golden Rules. But unless you are willing to pay between 50,000 or 400,000 Dollar per year (like most universities or wealthier companies) there is no way you can access these databases. The other more important reason is that these databases are designed to hinder research in the most effective way. Checking 1000 molecular formulae or structures for consistency or database coverage in one single step is impossible. The CAS DB SciFinder allows one formula search at a time via a manual search window. However the Seven Golden Rules need to check ten-thousands of formulae in one step. Also the export of obtained results is limited, either by exporting structures as GIF pictures instead of chemical structure codes (like CAS) or by limiting export to 200 results like in the case of Beilstein; hence hindering any useful way of research. Also these databases have no external web-connections to other databases or services, like the famous CACTVS DB, hence they are dead-end streets.
However to be very clear - to blame are not ACS (CAS provider) or MDL (Beilstein owner) with hundreds of scientist and curators doing their best job. To blame are hundreds of thousands of chemists and biologists worldwide and maybe even you (my valued reader) who refuse to deposit their molecules and meta-data in open-access archives or maybe even refuse to publish in open-source journals. Hence there is no opportunity for automated services (software robots) to search through these freely available datasets and built new innovative services (free or commercial) out of it.
Why annotation and not true idenfication?
The aim of metabolomics is the correct identification of a given structure via MS or NMR. However the number of unknown compounds is so large and the concentration range covers many orders of magnitudes so that a correct identification is not always possible. Therefore the annotation of a molecule with a name, similar to the peptide annotation score, is one possible solution. The name must be obtained by experiment and multiple matching factors (like matching retention time, mass spectrum, MS/MS spectrum or NMR spectrum). If multiple known structures are possible a metabolite score can be assigned.
Update on database developments
During the last two years there was a tremendous development in small molecule database and development of application programming interfaces (API). Allthough it is always possible to download the full PubChem data to calculate everything locally the curation and update process is a very cumbersome process, hence programmatic online access is preferred. Where PubChem is certainly the authority in small molecule databases, services like ChemSpider can provide streamlined or customized licensed solutions. API access should not be confused with general web or internet capability. API access means a program or tool or script can send information to a database and retrieve the answers sets back and can work with the results.
- PubChem PUG (Power User Gateway) - [LINK]
- ChemSpider API (including MS API) - [LINK] [MassSpec API]
- eMolecules (commercial chemicals) - [LINK]
- ChemNavigator (commercial chemicals) - [LINK]
- CSLS Beta (52 million structures) - [LINK]
- Soaring Bear (PhD) - A very complete database overview (and more)
- Sixty Four free databases - A comprehensive overview from depth-first (Rich Apodaca)