MS2Analyzer – software for substructure annotation of accurate mass MS/MS spectra
Project Partner: Yan Ma, Tobias Kind, Dawei Yang, Carlos Leon, Oliver Fiehn
MS2Analyzer – a software for small molecule substructure elucidation from accurate mass MS/MS spectra
Mass spectral features such as specific neutral losses and product ions can be used for substructure annotations, especially when reference libraries are not available. We developed a Java program named MS2Analyzer to search neutral losses, product ions, m/z differences and precursor ions in thousands of MS/MS spectra. It is designed for general small molecule substructure elucidation and can be applied to any specific study, such as lipid and glycoside identification.
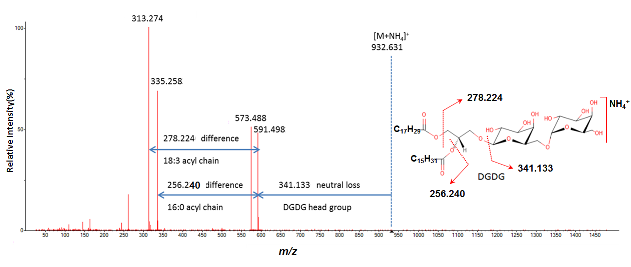
MS2Analyzer is a Java-based program for the analysis of mass spectral features such as neutral losses and product ions differences and product ions itself to allow for substructure annotations. The input file is the popular MGF format containing MS/MS spectra and the output is presented as EXCEL table that can be opened in Microsoft EXCEL or OpenOffice Calc for further statistical analysis. MS2Analyzer also works for MS1 data, in such a case the MGF file has to be created from MS1 scans only. This can be useful for complex sugar, saponins or polyflavonoid analysis.
Software download: The software is freely provided for commercial and non-commercial use under the MIT (http://opensource.org/licenses/MIT) license. The program and source code and examples can be downloaded here.
1. MS2Analyzer User Manual [PDF] (PDF file, 756 KB)
2. MS2Aanlyzer lipid annotation tutorial [PDF] (PDF file, 1.91 MB)
3. MS2Analyzer JAVA JAR executable program [ZIP] [ZIP2]
Related information:
Any question? Please feel free to email me: This email address is being protected from spambots. You need JavaScript enabled to view it.